Sampling AMR data, storing, and rendering it
The goal of this article is compare storage formats and rendering of Finite Element solutions produced with ASPECT. The computational mesh in ASPECT is a collection of adaptively refined octrees in 3d, see an example image below. For this experiment, we consider a stationary test benchmark in a unit cube that is part of ASPECT, see nsinker.
As part of this NSF funded project, we are evaluating how sampling unstructured data to a structured mesh works, and how storage and rendering of this structured data compares to the original unstructured output produced by ASPECT and the underlying deal.II library.
Parallel sampling of unstructured AMR data
For this experiment, we wrote an ASPECT postprocessor that sampled arbitrary solution variables on an unstructured AMR grid to a structured grid of arbitrary resolution. The postprocesor can be run at different resolutions to produce “multi-resolution” output.
The structured data is generated by looping over all cells, evaluating the solution variables at each quadrature point for some quadrature rule (slower to evaluate but more accurate with more quadrature points per cell). We then use nearest neighbor interpolation (the values at the closest quadrature point are used for each structured grid point). See the following image for an example:
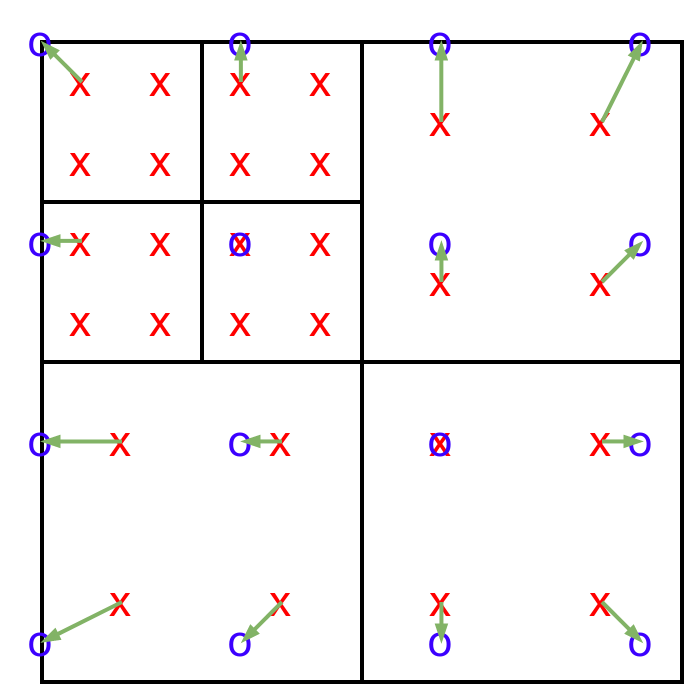
Here, black is the unstructured mesh, red are the quadrature points in each cell, blue circles are the points of the structured mesh, and the arrows denote what data is used at each point. Notice that a more sophisticated interpolation could be used, but this is certainly accurate enough for graphical visualization.
Internally, the algorithm transforms each quadrature point location to index space and then “splats” the solution to close by structured points. We keep track of the real world distance to the currently closest quadrature point in each structured point (as an additional output variable). When “splatting”, we only overwrite the current value, if the new distance is smaller than the stored one.
This also works in an MPI parallel computation, because we can split the structured mesh between processors and we know based on a given index, who the owner is. Each rank sends a list of indices with values and their distance to the owner, who then performs a similar “splat” operation like it is done for the own values.
The structured mesh is then output in parallel using the netCDF library with the HDF5 backend.
Structured netCDF vs unstructured, compressed vtu
We now compare the output of a structured netCDF file against the unstructured VTU output (as done using deal.II). The example is done on a sequence of adaptively refined meshes (based on the viscosity, visualized below). We output 6 data values in double precision (this corresponds to x,y, and z velocity, pressure, temperature, and viscosity here.
The unstructured solution in one of the intermediate steps looks like this (we show surface rendering, volume rendering, and isosurface rendering with 3 contours):
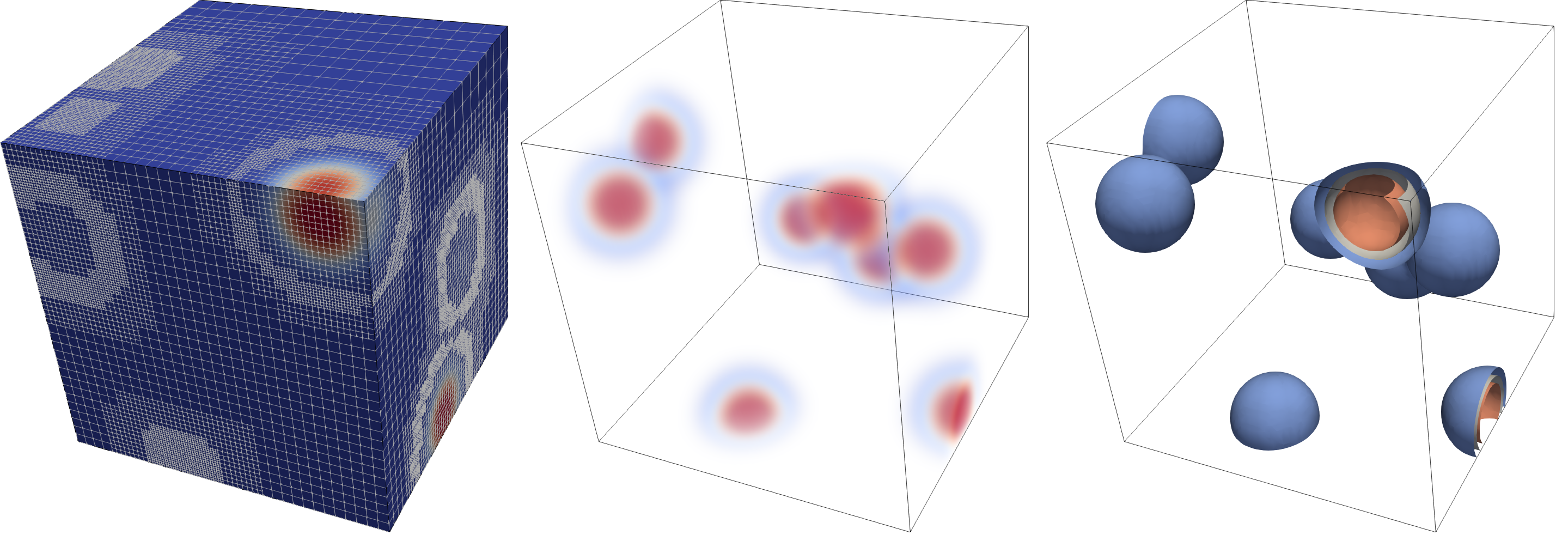
The unstructured data stores an unstructured list of cells (the leaves of the octree) with vertex coordinates in an compressed VTU file format. This is done by sending the binary representation of the data through zlib, followed by a base64 encoding to end up with a valid XML VTU file.
The structured data at various resolutions looks like this:
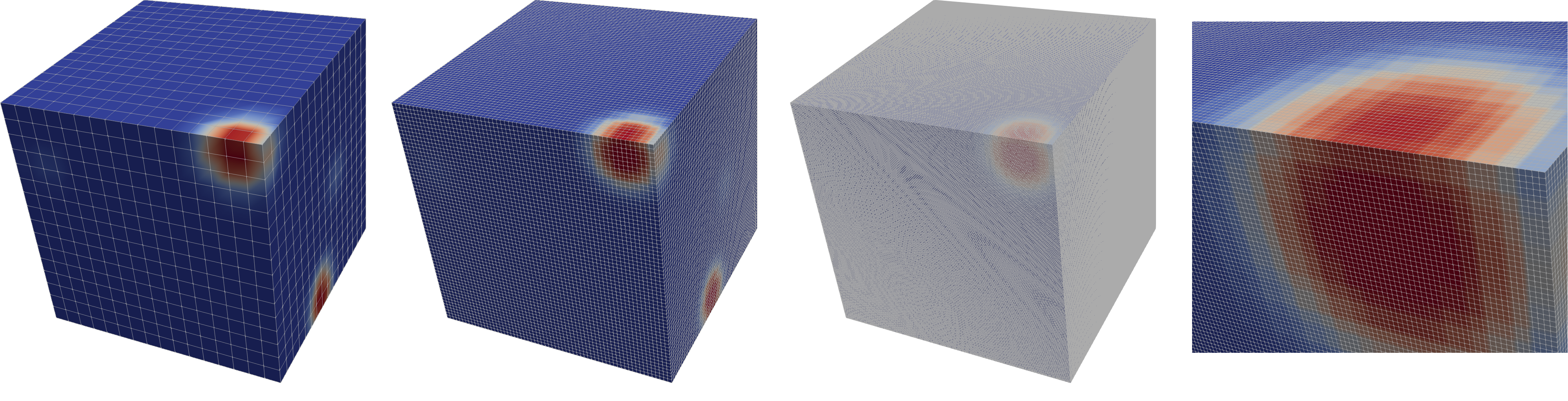
The netCDF file stores the data as binary using HDF5 directly and without compression (also see below for compression).
Now to the results. We start with the structured data:
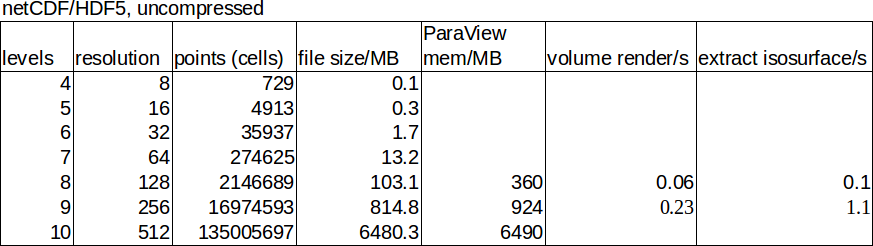
The table show the resolution, file size, and memory consumption and render time inside ParaView for selected data points.
For comparison, this is the unstructured data:
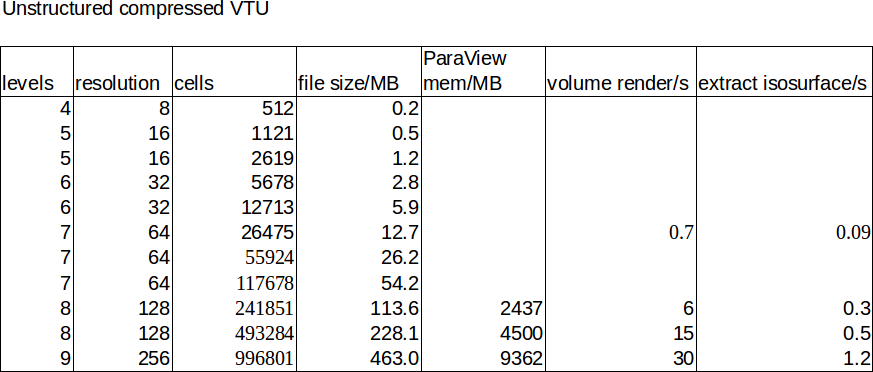
Here, the resolution refers to the maximum resolution (compared to the table above). Notice that file size are orders of magnitude smaller, while rendering time and memory consumption inside ParaView are orders of magnitude slower.
To conclude:
First, contour rendering is very fast, regardless of method and resolution (0.01s not shown in the table), while contour extraction is quite a lot slower for an unstructured mesh.
Second, surface rendering is very fast for both methods (interactive framerates even on a laptop without GPU rendering).
Third, file sizes for structured meshes quickly become very large but memory consumption inside ParaView is very efficient.
float vs double netCDF/hdf5
An obvious question is if we can reduce the file size of the netCDF files. First, let’s consider storing the data as floats instead of doubles. The loss in accuracy is unlikely to be problematic for visualization, but as expected, we save a factor of 2 in file size. Rendering performance and memory consumption also improves by a similar factor:
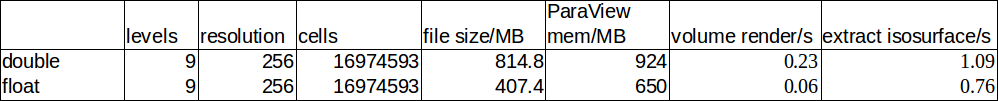
Other options
netCDF (HDF5) supports compression of the data (using nc_def_var_deflate()),
but it turns out that is not supported when writing data in parallel. This
makes it unusable for us during a computation. We could compress after the
fact using nccopy, as reading from compressed data should work in parallel.
See https://www.unidata.ucar.edu/blogs/developer/en/entry/netcdf_compression
for more details.
Conclusions
This little experiment showed some interesting results:
-
Structured data is a lot more efficient for rendering (RAM consumption, rendering time, contour extraction time)
-
High resolution structured data has very large file sizes (floats are a good option, but files will still be quite large without additional compression). Outputting at a lower resolution is certainly an attractive option as we are sampling the data already anyways.
-
All parts of the algorithms and the file sizes scale with the resolution, making it cheaper to produce the output as well. This makes structured, lower resolution output attractive as a data exchange format and to do quick visualization.
-
Sampling to structured data and compressing the data could be done as a postprocessing step in a separate code base after the fact if we store the unstructured data.
Future work
-
Clean up the sampling process and merge into ASPECT.
-
Update the code to support spherical geometries (lat/long/depth).
-
Compare unstructured data against vtkHyperOctree or vtkHyperTreeGrid from VTK. This requires investigation of the file formats.
-
Compare against OSPRay AMR rendering using TAMR.